Tutorials
Here are some additional recommended tutorials for metabarcoding data analysis
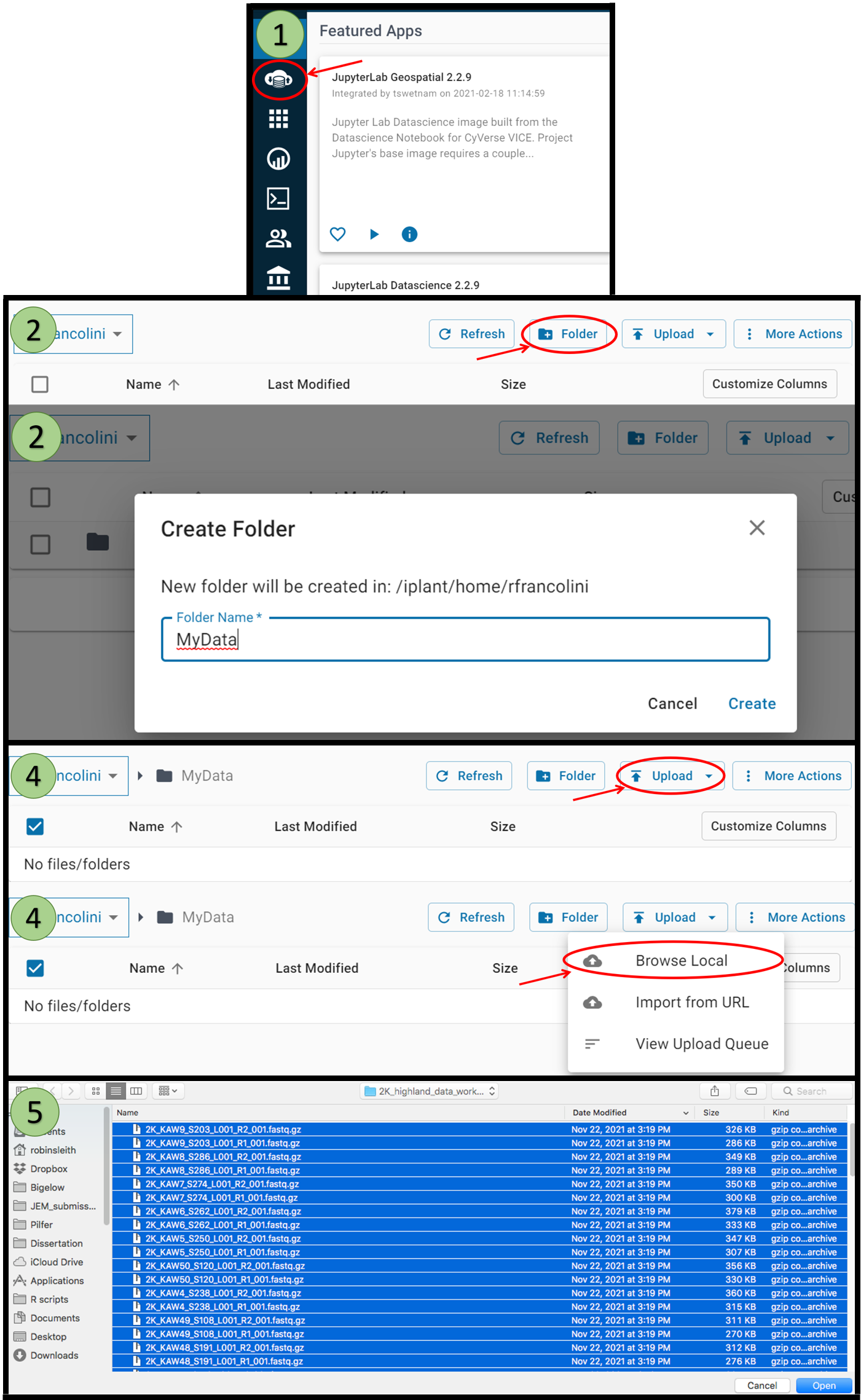
Uploading Data onto CyVerse
If you are interested in continuing working within the CyVerse environment with your own data, follow the instructions below to upload your data onto the server:
- Choose the data panel on the left
- Click the Folder button to create a new folder that will hold your reads
- Navigate to the newly created folder
- Click Upload then choose “Browse Local”
- Select files on your computer to be uploaded then choose “Open”
- Files will start uploading (this can take time if there are large files)
Publications
Callahan et al., 2016. DADA2: High-resolution sample inference from Illumina amplicon data. Nature Methods 13: 581-583.
Deiner et al., 2017. Environmental DNA metabarcoding: Transforming how we survey animal and plant communities. Molecular Ecology 26(21): 5872-5895.
Kaehler et al., 2019. Species abundance information improves sequence taxonomy classification accuracy. Nature Communications 10: 4643.
Liu et al., 2019. A practical guide to DNA metabarcoding for entomological ecologists. Ecological Entomology 45(3): 373-385.
Robeson II et al., 2021. RESCRIPt: Reproducible sequence taxonomy reference database management. PLoS Computational Biology 17(11): e1009581.
Ruppert et al., 2019. Past, present, and future perspectives of environmental DNA (eDNA) metabarcoding: A systematic review in methods, monitoring, and applications of global eDNA. Global Ecology and Conservation 17: e00547.
